NRPS/PKS modules#
Added in antiSMASH 5.1
NRPS and PKS products come from assembly lines of modules.
These modules are detected by antiSMASH and will be shown in the NRPS/PKS domains tab for a region, along with the domains from previous versions.
Here is an example of a single gene with multiple modules:

These modules shown are complete, in that they have a loader domain (e.g. adenylation or acyltransferase) and a carrier protein (e.g. ACP).
Complete modules may also have a starter domain (e.g. condensation or ketosynthase).
In the case of a trans-AT-PKS module, provided it has a trans-AT specific ketosynthase,
it will have no loader domain within the module, and the loaded substrate will be assumed to be Malonyl-CoA.
At the top of the section is a button Show module domains that will hide the module graphic and show the domains similarly to previous versions of antiSMASH.
The module borders and backgrounds will still be shown to indicate the extents of the module.
The same example from above is shown below with this domain option enabled.

The thioesterase (TE) domain shown is included in the final module.
It is not hidden by the module graphic due to being important for a whole product.
Moving the mouse cursor over a complete module will hide the module graphic and display the component domains. These domains can be clicked on to access specific information (this behaviour has not changed with the addition of modules).
Cross-gene modules#
Some modules can be split across two genes. These modules, when detected, are shown with jagged edges on the joining side. To help with identifying the other part of the module in the other gene, each part of the module is also labelled with a character.
Here is an example of these modules, using a small section of the kirromycin gene cluster:

Modifications#
The second module in the above examples has a modification domain (nMT or N-methyltransferase).
Modification domains contained by a module will modify the loaded substrate, with the modified substrate (or monomer), being displayed on the module.
In the examples above, the adenylation domain (A) is predicted to load leucine (leu), and is then modified by the module to be a N-methylated leucine (NMe-leu).
Incomplete modules and incomplete predictions#
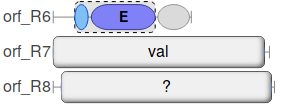
Along with complete modules, e.g. in orf_R7 above, some incomplete modules are annotated (e.g. orf_R6)
It is common to have large fragments of domains and multiple domains that by themselves don't complete a module.
In cases where these domains are found and cannot form a complete module, an incomplete module will be formed.
Incomplete modules are shown with a dashed outline and will always show component domains.
In the case of a loader domain without a carrier protein, the predicted substrate that would be loaded is not considered for a candidate cluster's polymer prediction.
There are also occasions, even in a complete module, where no consensus can be found for the prediction of a loaded substrate.
In these cases, the module will still be shown, but the substrate will be shown as ? as shown for orf_R8 in the image above.
Complete modules with unknown substrates will still apply modifications, thus monomers like D-NMe-? are possible.
For the purposes of a candidate cluster's polymer, these unknown predictions will use the generic nrp/X SMILES strings.
Iterative polyketides#
In the case of a PKS_KS domain being iterative, the module will show an additional icon. For example:

The number of iterations is not determined, and only a single monomer (in this case mal) will be added to the candidate cluster's final polymer.
Bubble/module-only drawing#
An alternative visualisation of modules was added after the release of antiSMASH 6.0.
This visualisation will be familiar to those who are reading academic papers of related cluster types.
This is accessed by selecting the NRPS/PKS modules details tab in the HTML results.
For example, a small section of the kirromycin cluster:
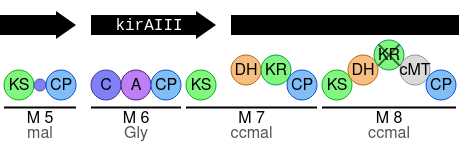
In this visualistion, the gene ordering is as predicted for that particular candidate cluster (which can be changed in a drop-down menu above the drawing itself).
Gene arrows, which can be clicked to highlight that gene in the cluster viewer, indicate the extent of genes.
Modules are numbered, e.g. M 5, and shown with lines that indicate the extent of the module.
In the example above, module 7 is a cross-gene module, with the module line crossing the boundaries of kirAIII and the gene to the right.
Below the module number is the predicted monomer of the module.
A legend is included in the visualisation to explain the various notations, one example of which is the inactive KR domain in module 8.