Transcription Factor Binding Site (TFBS) Prediction#
Transcription factor binding sites are predicted using the LogoMotif profiles.

On the region overview, predicted TFBS hits are shown with small black pins.
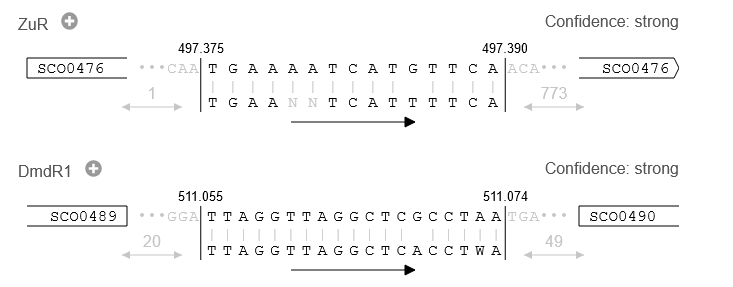
In the "TFBS Finder" tab in the details panel, the different TFBS hit are shown with their binding site sequence and the surrounding genes for context.
The "TFBS Finder" sidepanel contains an overview of all the TFBS hits found in the region.
Database#
| Site name | Description | Source organism |
|---|---|---|
| ANR | Anaerobic transcriptional regulator | Pseudomonas putida KT2440 |
| AbrC3 | Antibiotic production activator | Streptomyces coelicolor |
| AfsQ1 | Two-component system AfsQ1-Q2, activator of antibiotic production | Streptomyces coelicolor |
| AfsR | Pleiotropic regulatory for antibiotic production | Streptomyces coelicolor |
| ArcA | Repressor of carbon oxidation | Escherichia coli str. K-12 substr. MG1655 |
| ArgR | Regulator of arginine biosynthesis genes | Streptomyces coelicolor |
| BldD | Development and antibiotic global regulator | Streptomyces coelicolor |
| CRP | Cyclic AMP receptor protein | Escherichia coli str. K-12 substr. MG1655 |
| CatR | H2O2-responsive repressor | Streptomyces coelicolor |
| CcpA | Catabolite control protein A | Streptococcus & Clostridium sp. |
| CelR | Cellobiose uptake repressor | Streptomyces coelicolor |
| ClgR | Caseinolytic protease gene regulator | Corynebacterium glutamicum ATCC 13032 |
| CodY | Regulator of stationary phase and virulence in Gram-positive bacteria | Bacillus, Lactococcus & Streptococcus sp. |
| ColR | Phenol-responsive regulator | Pseudomonas putida KT2440 |
| CopR | Copper-responsive regulator | Lactococcus lactis subsp. lactis Il1403 |
| CsoR | Copper-responsive repressor | Streptomyces lividans |
| DasR | N-acetylglucosamine dependent repressor | Streptomyces coelicolor |
| DmdR1 | Iron(II)-dependent repressor | Streptomyces coelicolor |
| DosR | Hypoxia stress response regulator | Mycobacterium tuberculosis H37Rv |
| FuR_variant_1 | Ferric Uptake Regulator | Bacillus, Pseudomonas, Yersinia sp. |
| FuR_variant_2 | Ferric Uptake Regulator | Caulobacter crescentus CB15 |
| GnfM | Regulator of nitrogen-fixation genes | Geobacter sulfurreducens PCA |
| HexR | Glucose-responsive regulator | Shewanella oneidensis MR-1 |
| HgtR | Hydrogen-dependent regulator | Geobacter sulfurreducens PCA |
| HipB | Bacterial antitoxin HipB | Escherichia coli BL21(DE3) |
| HypR | L-hydroxyproline utilization repressor | Streptomyces coelicolor |
| IolR | Regulator of the inositol catabolism | Caulobacter crescentus CB15 |
| LexA_variant_1 | Repressor of DNA damage response | Streptomyces coelicolor |
| LexA_variant_2 | Repressor of DNA damage response | Bacillus subtilis subsp. subtilis str. 168 |
| LexA_variant_3 | Repressor of DNA damage response | Escherichia coli str. K-12 substr. MG1655 |
| LuxO | Repressor of luminescence | Vibrio campbellii ATCC BAA-1116 |
| MatP | Cell cycle regulator | Escherichia coli str. K-12 substr. MG1655 |
| MexT | Global virulence regulator | Pseudomonas aeruginosa PAO1 |
| MntR | Manganese transport regulator | Escherichia coli str. K-12 substr. MG1655 |
| MogR | Repressor of flagellar motility genes | Listeria monocytogenes EGD-e |
| NrdR | Represses ribonucleotide reductase encoding genes | Streptomyces coelicolor |
| NrtR | NAD synthesis repressor | Streptomyces coelicolor |
| NtrC | Nitrogen regulatory protein C | Pseudomonas putida KT2440 |
| NuR | Nickel-responsive regulator | Streptomyces coelicolor |
| OsdR | Development and stress management regulator | Streptomyces coelicolor |
| PerR | Peroxide regulator | Bacillus & Staphylococcus sp. |
| PsrA | Oleic acid responsive regulator | Pseudomonas sp. |
| RicR | Copper-responsive regulator | Mycobacterium tuberculosis H37Rv |
| RpoN | Regulator of nitrogen assimilation and virulence | Salmonella & Vibrio sp. |
| RutR | Regulator of the pyrimidine and purine metabolism | Escherichia coli str. K-12 substr. MG1655 |
| SypG | Regulator of biofilm formation and host colonization | Vibrio fischeri ES114 |
| ToxT | Regulator of MSHA biosynthesis | Vibrio cholerae O395 |
| VqsM | Regulator of quorum-sensing signalling systems | Pseudomonas aeruginosa PAO1 |
| ZuR_variant_1 | Zinc-responsive repressor | Streptomyces coelicolor |
| ZuR_variant_2 | Zinc-responsive repressor | Pseudomonas protegens Pf-5 |