CompaRiPPson#
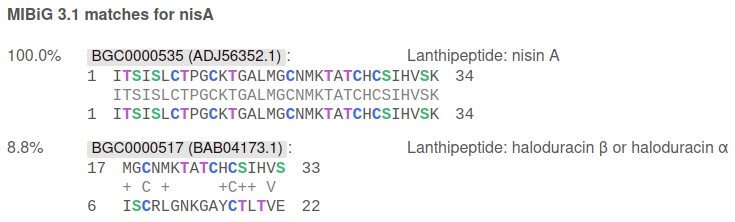
Ribosomally synthesised and post-translationally modified peptide (RiPPs) precursors are compared to various databases. An example of results from the MIBiG database for the nisin A precursor is below:
Database details#
The general format applies to all databases, with the database name and version displayed above the hits for that precursor and that database.
In the example above, that is MIBiG 3.1, for the precursor nisA.
Hit details#
Following the database information are the details of the best hits, with weaker hits initially hidden.
The first line of each hit starts with the identity percentage of the hit and the precursor. This is followed by the accession of the database entry (grouped together in instances where the precursor sequence is shared). The last item on the line is a description of the hit, beginning with the type of RiPP. For MIBiG hits, the compound(s) produced by the reference cluster follows the RiPP type. For other databases such as the antiSMASH database, the locus tag of the precursor gene is shown.
After the descriptive information is an alignment of the precursor translation and the reference translation. The query is shown as the top line, and the reference is the bottom line, with the mid line being the consensus. Numeric values to the left are the start position (within the translation) of the match, and the end position to the right.
An example of results from the antiSMASH-DB is shown below, including a shared reference sequence:
